Make your ECL more dashing using the dapper bundle
At Proagrica we’ve been using HPCC Systems for our data storage and transformation for a couple of years. We’ve learnt a lot over this time but there’s something that comes up quite frequently while we are testing, doing quick investigations or making some model features. The issue is that ECL can be a bit verbose and sometimes simple things are not simple to remember. It can be especially challenging among occasional users who may need to lookup things whenever they open the ECL IDE.
We felt this pain and so developed an ECL bundle to make coding faster, more logical and most of all, easier to remember. This package has become known as dapper and it allows us to quickly subset, transform and summarise data. If you’ve used R’s dplyr package you may well get what we mean!
Intrigued? Me too. Let’s look at what it can do!
Dapper is available as an ECL bundle. Something that we only recently learned is that HPCC Systems supports libraries in a similar way to Python’s pip. The basic idea is that it will pull down a ‘bundle’ of ECL code from github and install it locally making it available as if it was part of the core libraries.
So you can IMPORT a bundle without having to have the scripts in your repo.
The dapper tool is made available with ecl.exe (and its Linux equivalent). To install the latest dapper release you simply run the following on the command line (although I hear that the latest ECL IDE has this baked in).
ecl bundle install -v https://github.com/OdinProAgrica/dapper.git
There are many more ECL bundles available. View the full list in the HPCC Systems ECL Bundles github repository.
What can dapper do?
Once installed, dapper can be used to create scripts using simple verbs which can increase readability and decrease coding mistakes. In short, it reduces the amount of time you spend thinking about how to do a job, you can just get on with it.
Don’t get me wrong, sometimes an old school PROJECT is better. I’ll leave it to you to make that decision. As always, the right tools are needed for the right job. However, it is worth noting that even if you use several TransformTools statements in a row, the compiler is clever enough to combine this into a single operation under the bonnet, minimising dapper’s speed impact.
The bundle itself is broken down into two sets of tools. This post will focus on our transform tools. I will do a separate post on stringtools (and our in development bundle geodapper) in the future. Watch this space!
IMPORTing dapper
Please note: The standard nomenclature for importing the modules should be respected.
This is because each module references its own functions requiring a known import name. Modules should always be imported as:
- TransformTools: tt
- StringTools: st
For example:
Import dapper.TransformTools as tt;
Import dapper.StringTools as st;
Also note that inDS and OutDS are reserved! You may find you get weird errors if you use these variable names, this will be fixed in a later release.
Transform Tools
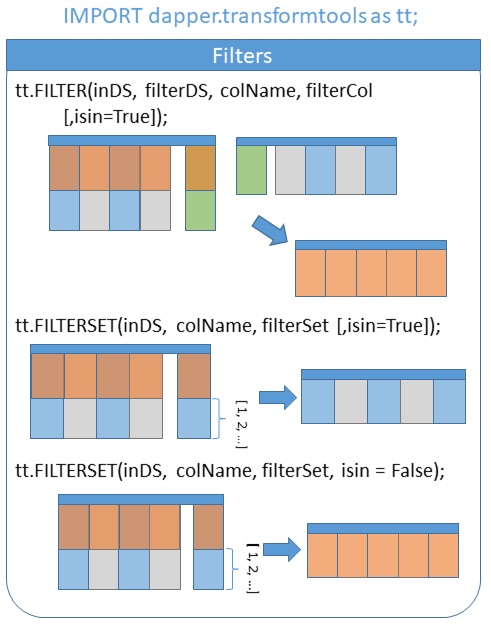
Have a quick skim over dappper’s key functions below. I’ve just chosen a subset here, a more complete list is available on the project’s github and includes things like filter operations. Once you’ve got the gist I’ll give you an ECL script we can pick apart.
Data Transformations
Duplicates
Column Transforms
Filters
Arrangement
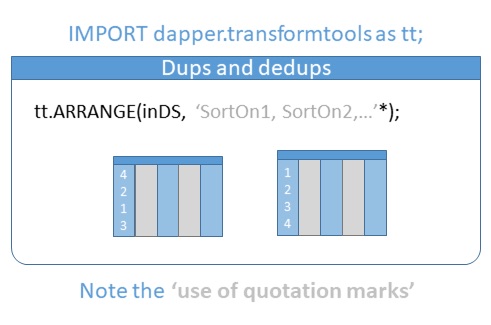
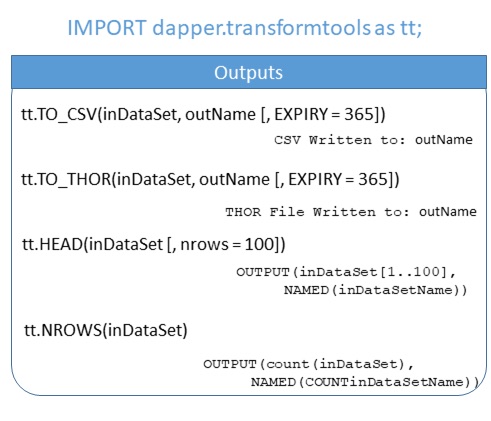
Outputs
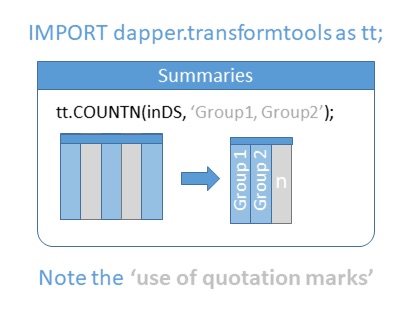
Summaries
How does dapper work?
Here is an ECL script which (assuming you have installed dapper using the bundle method described above) will work on your system.
IMPORT dapper.ExampleData;
IMPORT dapper.TransformTools as tt;
//load data
StarWars := ExampleData.starwars;
// Look at the data
tt.nrows(StarWars);
tt.head(StarWars);
//Fill blank species with unknown
fillblankHome := tt.mutate(StarWars, species, IF(species = '', 'Unkn.', species));
tt.head(fillblankHome);
//Create a BMI for each character
bmi := tt.append(fillblankHome, REAL, BMI, mass/height^2);
tt.head(bmi);
//Find the highest
sortedBMI := tt.arrange(bmi,'-bmi');
tt.head(sortedBMI);
//Jabba should probably go on a diet.
//How many of each species are there?
species := tt.countn(sortedBMI,'species');
sortedspecies := tt.arrange(species, '-n');
tt.head(sortedspecies);
//Finally let's look at unique hair/eye colour combinations:
colourData := tt.select(StarWars, 'hair_color, eye_color');
unqiueColours := tt.distinct(colourData, 'hair_color, eye_color');
//see arrangedistinct() for fancy sort/dedup operations.
tt.head(unqiueColours);
tt.countn(unqiueColours);
//and save our results
tt.to_csv(sortedBMI, 'ROB::TEMP::STARWARSCSV');
Let’s break this down
Example Dataset
IMPORT dapper.ExampleData; IMPORT dapper.TransformTools as tt; //load data StarWars :=ExampleData.starwars;
Viewing Data
// Look at the data tt.nrows(StarWars); tt.head(StarWars);
These are shorthand for OUTPUT and COUNT, however note the way the results are named. It automatically renames your results to match your variables.
Transformations
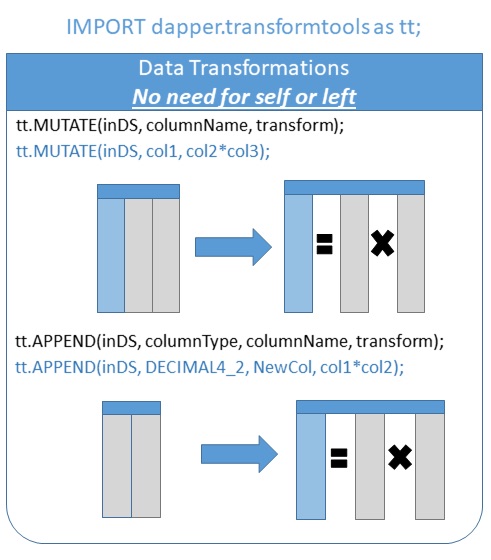
//Fill blank species with unknown fillblankHome := tt.mutate(StarWars, species, IF(species = '', 'Unkn.', species)); tt.head(fillblankHome); //Create a BMI for each character bmi := tt.append(fillblankHome, REAL, BMI, mass/height^2); tt.head(bmi); //Find the highest sortedBMI := tt.arrange(bmi, '-bmi'); tt.head(sortedBMI); //Jabba should probably go on a diet.
Mutate and append allow column transforms via a simple formula. Note that there is no need for SELF or LEFT in these transforms, making them easier to write and easier to read!
Grouped Counts
//How many of each species are there? species := tt.countn(sortedBMI, 'species'); sortedspecies := tt.arrange(species, '-n'); tt.head(sortedspecies);
The record definition to do a cross-tab is something I always have to look up. countn will do it for you, you can even hand multiple columns to it and it’ll handle them all perfectly.
Deduplication and Column Selection
//Finally let's look at unique hair/eye colour combinations: colourData := tt.select(StarWars, 'hair_color, eye_color'); uniqueColours := tt.distinct(colourData, 'hair_color, eye_color'); tt.head(uniqueColours); tt.countn(uniqueColours);
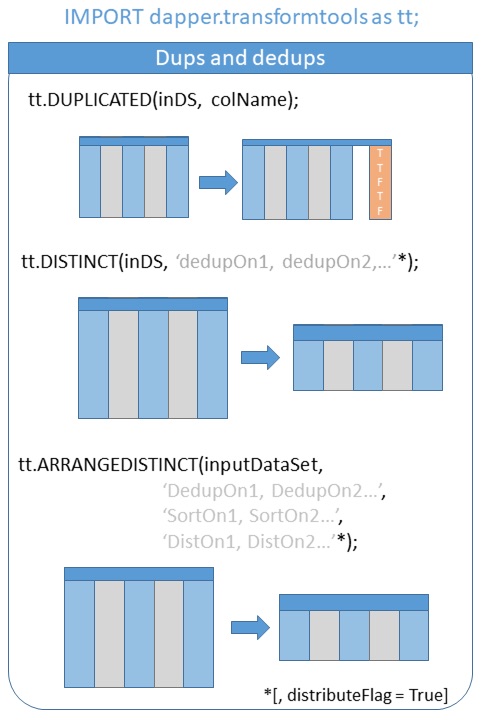
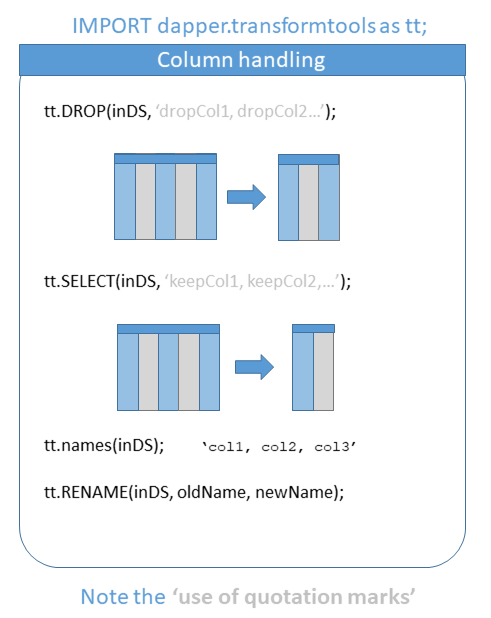
‘select’ (and it’s partner function ‘drop’) will help in quickly sub-setting data. Note also the use of ‘distinct’ which is shorthand for DEDUP(SORT(DISTRIBUTE(…), LOCAL), LOCAL).
Using arrangedistinct() instead allows you to control the sort and distribute commands separately. See also ‘duplicated’ which will flag all duplicates with a boolean for investigation.
Saving Results
//and save our results tt.to_csv(sortedBMI, 'ROB::TEMP::STARWARSCSV');
Finally we can write out.
OUTPUT(…CSV(…)) is another one of those functions that often gets forgotten. It handles all the quote, separator, header stuff for you and simply writes out a ‘normal’ csv file. It also adds the tilde (~) to the start of your file name if it’s not already there.
How would this look in ECL?
Just in case your interested…
IMPORT dapper.ExampleData;
//load data
StarWars := ExampleData.starwars;
// Look at the data
OUTPUT(COUNT(StarWars), NAMED('COUNTstarWars'));
OUTPUT(StarWars, NAMED('starWars'));
//Fill blank species with unknown
//Create a BMI for each character
fillblankHomeAndBMI := PROJECT(StarWars, TRANSFORM({RECORDOF(LEFT); REAL BMI;},
SELF.BMI := LEFT.mass / LEFT.Height^2;
SELF.species := IF(LEFT.species = '', 'Unkn.', LEFT.species);
SELF := LEFT;));
OUTPUT(fillblankHomeAndBMI, NAMED('fillblankHomeAndBMI'));
//Find the highest
sortedBMI := SORT(fillblankHomeAndBMI, -bmi);
OUTPUT(sortedBMI, NAMED('sortedBMI'));
//Jabba should probably go on a diet.
//How many of each species are there?
CountRec := RECORD
STRING Species := sortedBMI.species;
INTEGER n := COUNT(GROUP);
END;
species := TABLE(sortedBMI, CountRec, species);
sortedspecies := SORT(species, -n);
OUTPUT(sortedspecies, NAMED('sortedspecies'));
//Finally let's look at unique hair/eye colour combinations:
colourData := TABLE(sortedBMI, {hair_color, eye_color});
unqiueColours := DEDUP(SORT(DISTRIBUTE(colourData, HASH(hair_color)), hair_color, eye_color, LOCAL), hair_color, eye_color, LOCAL);
OUTPUT(COUNT(unqiueColours), NAMED('COUNTunqiueColours'));
OUTPUT(unqiueColours, NAMED('unqiueColours'));
//and save our results
OUTPUT(sortedBMI, , 'ROB::TEMP::STARWARSCSV', CSV(HEADING(SINGLE), SEPARATOR(','), TERMINATOR('\n'), QUOTE('"')));
Summary
We hope you enjoyed your quick whistle stop tour of dapper’s power and functionality. It is incredibly useful for testing and investigative work, allowing a more logical flow and readable code. If you do have any comments or suggestions please checkout the github issues page here. Contributions, as always, are also welcome.
For more information about how to create an ECL bundle see our ECL Bundles Writers Guide.
