Integrating Prior Knowledge with Learning in Biomedical Natural Language Processing
Natural language processing (NLP) research has progressed rapidly in the last year, with significant performance improvement in a variety of NLP tasks. Yet, the research of NLP techniques in the biomedical domain has not progressed much. The advantage of the biomedical domain is that it has a high-quality human curated knowledge base and large-scale literature, both of which contain rich prior knowledge.
improvement in a variety of NLP tasks. Yet, the research of NLP techniques in the biomedical domain has not progressed much. The advantage of the biomedical domain is that it has a high-quality human curated knowledge base and large-scale literature, both of which contain rich prior knowledge.
Jingqing Zhang, a PhD candidate at the Data Science Institute, Imperial College London, introduced his work, “Integrating Prior Knowledge with Learning in Biomedical Natural Language Processing” at HPCC Systems Tech Talk 33. The full recording of Jingqing’s Tech Talk is available on YouTube.
In this blog, we will look at Jingqing’s recent research on biomedical NLP, specifically the integration of prior knowledge with deep learning models.
Deep Learning with Prior Knowledge
Prior knowledge, in general, is common sense or background knowledge. When learning languages, it takes a long time to accumulate the experience, knowledge, and skills necessary to perform complicated communication tasks, like communicating with colleagues or doing a presentation.
When developing data models, it is important to consider background or prior knowledge in addition to future exceptions or statistical analysis of the data itself, so that more precise decisions can be made.
In the graphic below, in the learning box, there is an interaction between “Perception” and “Prior Knowledge,” which is received from “Knowledge Providers.”
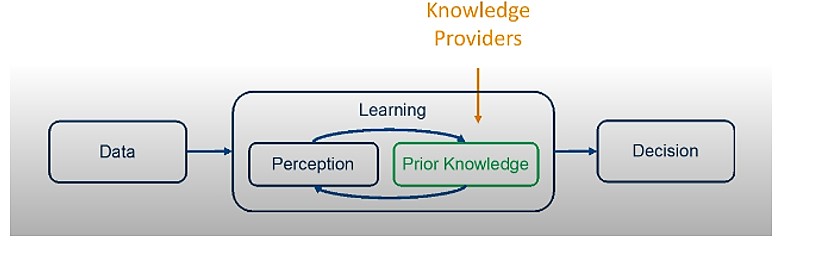
Prior Knowledge
There are two kinds of knowledge that are considered in Natural Language Processing:
Unstructured knowledge
- This type of knowledge be implicitly contained in large text corpus. In other words, unstructured knowledge can be obtained from a variety of sources, such as newspapers, books, blogs, etc.
- It is difficult to extract information from the text corpus and effectively use it for machine learning models. But, recently, there has been success using text corpus together with pre-training techniques to improve model performance.
Structured knowledge
- This type of knowledge can be explicitly defined by knowledge graphs, ontologies, or hierarchal trees.
- Structured knowledge is proven to be effective in tasks that require reasoning and understanding.
Motivation
The motivations for this work are that electronic health records (EHRs) are widely and increasingly being adopted, and there is a desire to understand the professional and natural languages in the medical domain with manually pre-defined biomedical ontologies.
The image below gives a visual overview of this research. The objective is to find the relationship between the plain text EHRs (written by clinicians), and the biomedical ontologies (pre-defined by biomedical experts), in order to provide structure for the EHRs. This can then be used for other purposes, such as analysis, diagnosis, treatment, medical chatbots, medical QA, etc.
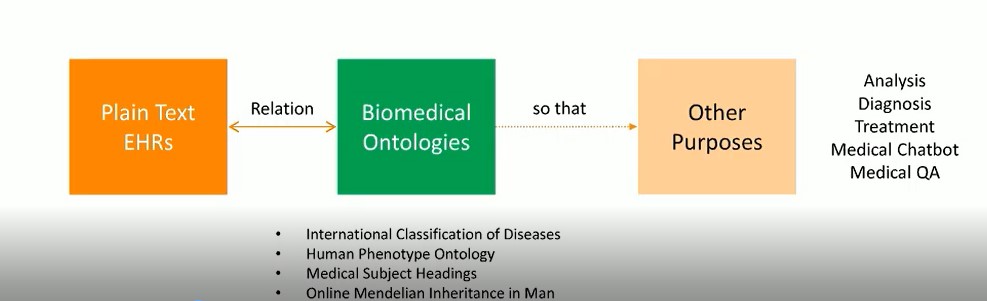
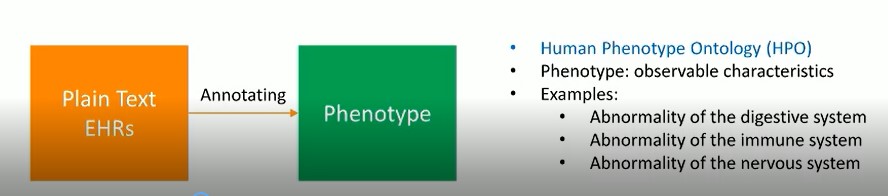
The goal of this research is to annotate EHRs with pre-defined phenotypes (deviations from normal morphology, physiology, or behavior). EHRs serve as a rich source of phenotype information. Patients diagnosed with the same disease can be further classified into sub-groups by phenotypes. Phenotype annotation of EHRs can provide more information for disease diagnosis, genomic diagnostics, and precision medicine.
Datasets
Experiments for this research were conducted using two datasets:
1. A public EHRs database (MIMIC-III) with 52,722 discharge summaries
- Each EHR also came with disease diagnosis marked by International Classification of Diseases (ICD-9) codes.

2. Human Phenotype Ontology (HPO) – Standardized knowledge base of HPO over 13,000 terms with descriptions.
- In HPO, each phenotypic abnormality term has a name, synonyms, and a definition.
- The HPO also provides the class-subclass relations between phenotypic abnormalities.

The EHRs were randomly split into a training set (70%) and a held-out set for testing (30%).
Research
For this work, the research team proposed a novel unsupervised deep learning framework to utilize supportive phenotype knowledge in HPO and annotate general phenotypes from EHRs semantically.
The research team demonstrated that the proposed method achieved state-of-the-art annotation performance and computational efficiency compared with other methods.
Problem Formulation (Example)
The following is example of what the annotation looks like:
The left side shows EHR tags describing the clinical characteristics of patients. The right side shows potential annotations with HPO terms.
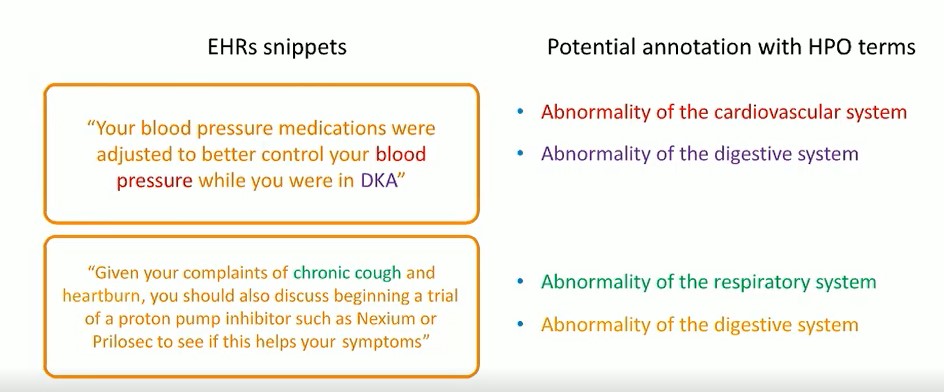
It is not easy to define all the rules to extract the phenotypes. There are many different expressions that can describe the same thing. For this reason, the semantic method was chosen. No matter how different the expression is, as long as the context can show this information, the phenotype information can be detached from the semantics.
There are two types of data sources.

For this research, the focus is only on the primary category of phenotype. The general phenotypic abnormalities (in the orange box) are what the model seeks to annotate from EHRs. Meanwhile, the HPO provides additional subclasses. The solid lines show direct relation and the dashed lines show relation with multiple hops. A HPO (red circle) can be a subclass of multiple Hj’s. The S(Hj) in the green box stands for the additional subclasses of Hj .
In the hierarchy, from the root node to the first level of phenotype, there are about 24 categories of phenotypes, and the focus is only on these categories. There are more detailed HPOs available below the general categories, and that is used as the point of information in the training of the model.
The EHR can include multiple, single, or no phenotypes. It is a question of learning the conditional probability versus whether a specific phenotype is mentioned inside an EHR. There is a probability of 0 or 1, with 1 meaning that the EHR mentions the phenotype and can be annotated, and 0 meaning that the phenotype has nothing to do with the EHR. The following is the equation for conditional probability:
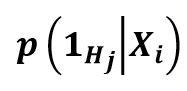
- I.e. a binary classification for each Hj
- As a whole, a multi-label classification on subclass H
Semantic Latent Representations
The methodology for this research is based on two assumptions:
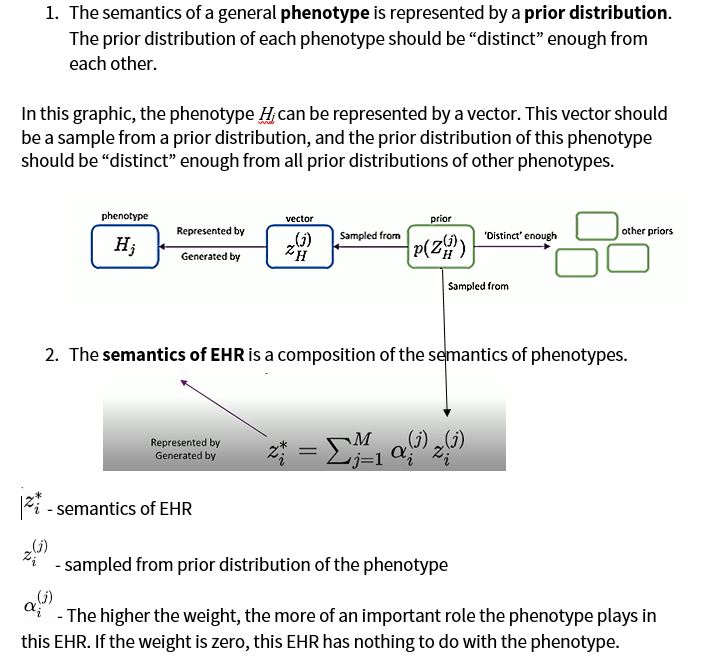
Before discussing the proposed model for this research, it is important to note that the textual data used was prepared using the HPCC Systems platform.
HPCC Systems
The textual data used in this research was processed using the HPCC Systems platform. A Python embed on ECL (Enterprise Control Language) was used for analysis.
An auto-encoder model

The model below shows the proposed deep learning framework for this research.
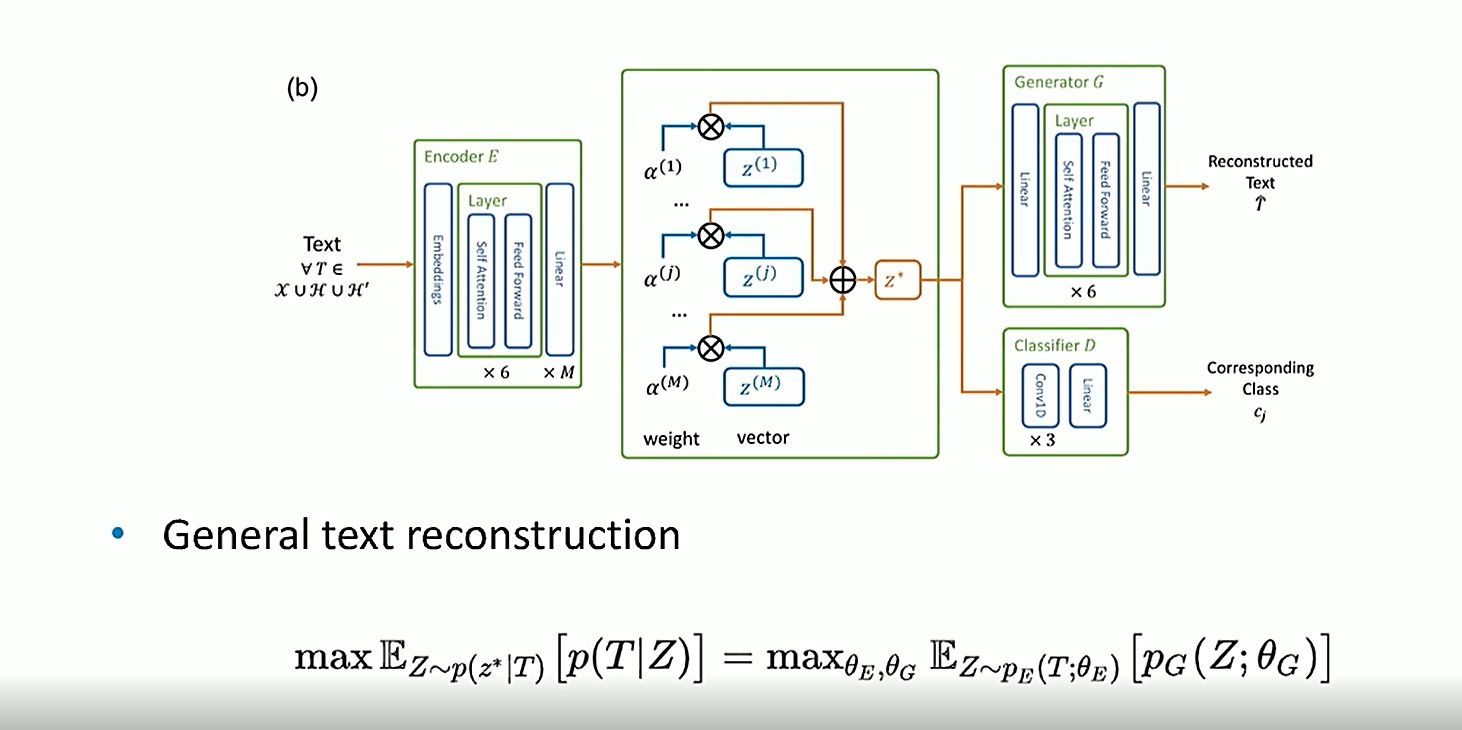
For general text reconstruction, texutal data enters the Encoder. It then flows to the semantics of EHR in the middle, where the weight and vector dimensions are applied, and then on to the weight of composition (z*). It then goes to the Generator G, where the textual data is reconstructed. The Classifier D (cj) is used to constrain and differentiate the priors.
The goal is to learn which phenotypic abnormalities are semantically more important in EHRs.
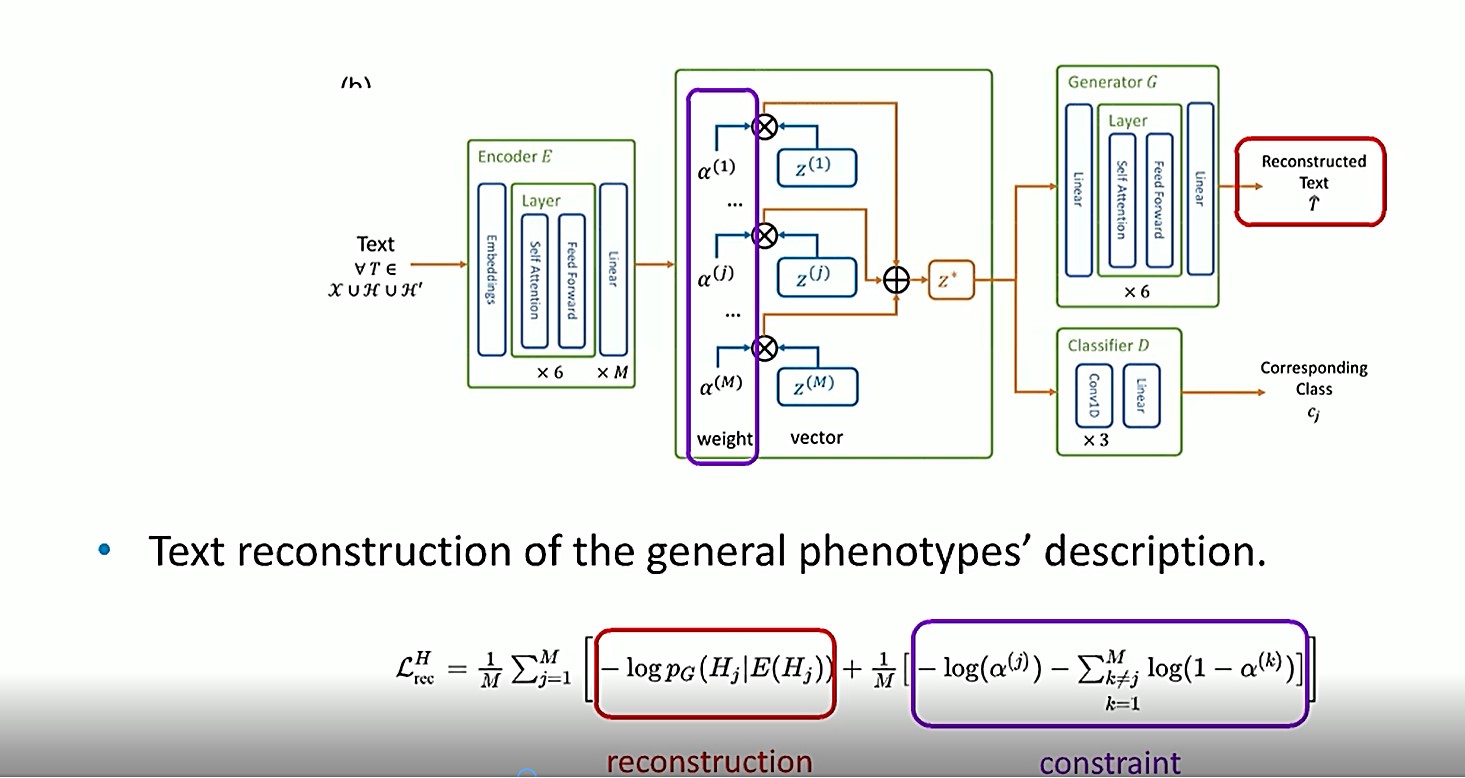
In the model shown above, an extra constraint is applied to train the reconstruction. For example, if the description of phenotype ”j” is given so that the corresponding weight, ?(j), is 1, or as high as possible, the weights of all the other phenotypes should be close to 0, because they have nothing to do with the reconstruction.
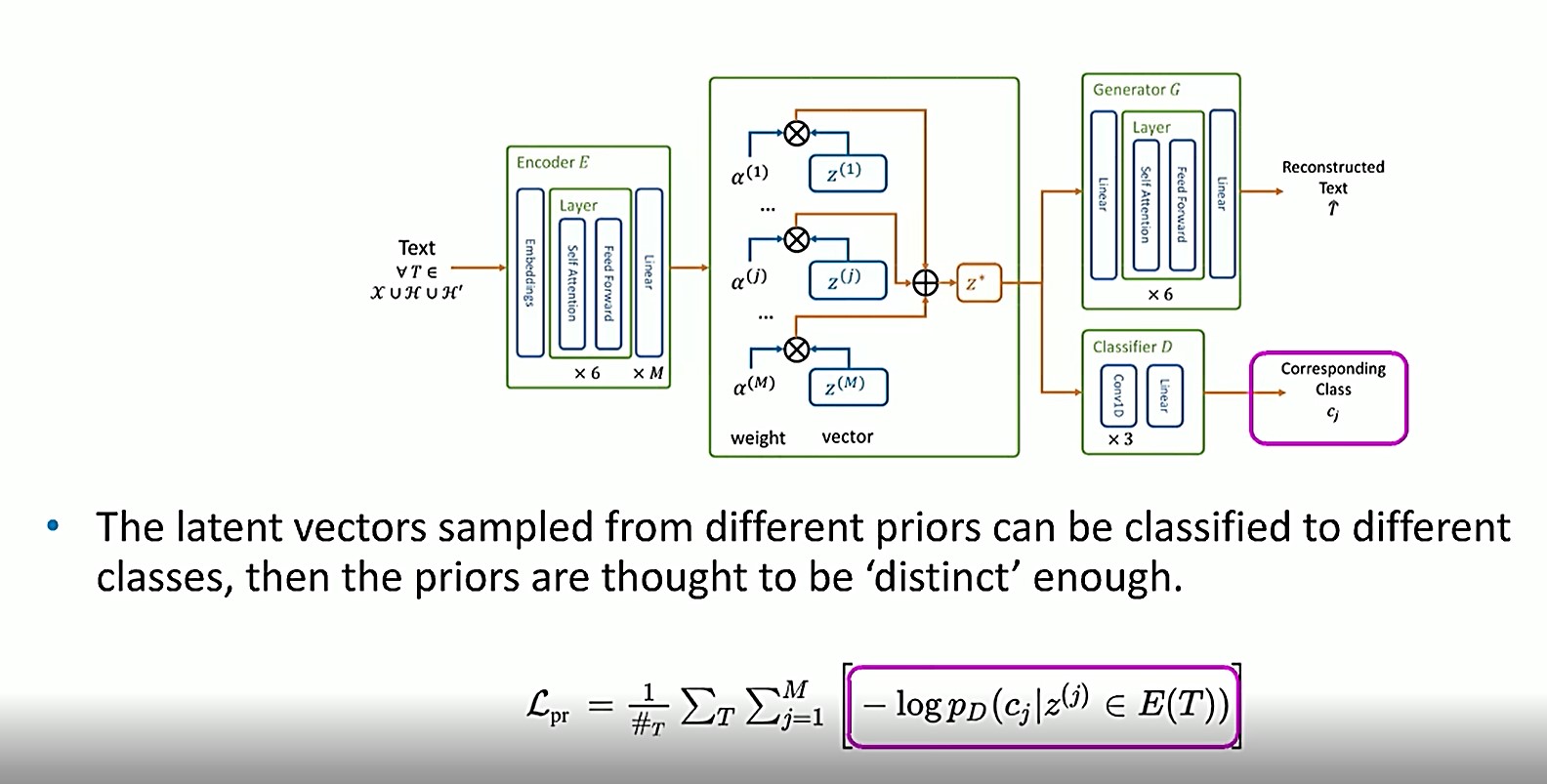
In this equation, another classifier is applied as a constraint to make sure that each of the vectors z(1), z(j), z(m) can be classified into the correct classes.
Experiments – Time Efficiency
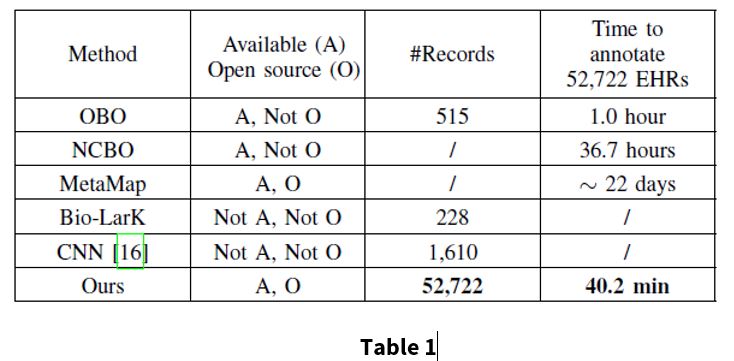
Table 1 compares different methods to show the scalability and efficiency of the proposed model.
In table 1, the #Records referes to the number of textual records used in the original works. The time was measured by the duration of annotating 52,722 EHRs in inference state with a single thread Intel I7-6850K 3.60GHz and and single NVIDIA Titan X.
Automatic annotation techniques based on natural language processing (NLP), that use indexing and retrieval techniques include:
- OBO Annotator
- NCBO Annotator
- MetaMap
- Biolark
Indexing and retrieval techniques require manually defined rules, and can be computationally inefficient.
CNN represents supervised deep learning models used to annotate phenotypes. These models are effective, but training is difficult.
For the proposed method, experiments were conducted on 52,722 EHRs, which is considerably more than previous works. The proposed method is also computationally more efficient than the baselines. The annotation stage of this method takes 40.2 minutes to complete, which is much faster than the other annotation methods, as seen in Table 1.
Experiments – Accuracy
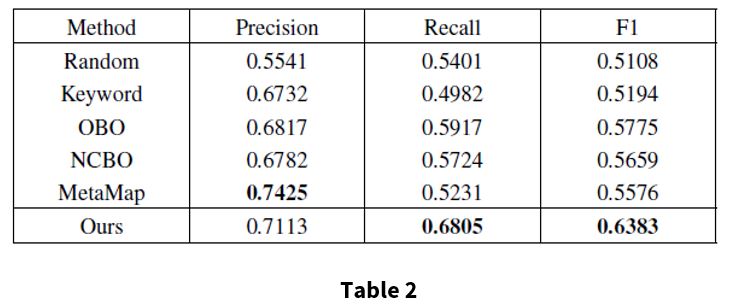
Table 2 shows the performance of annotation results compared with the silver standard. All numbers are averaged across EHRs in the testing set. The proposed method achieves a precision of 0.7113, recall of 0.6805, and F1 0f 0.6383. Based on these results, the proposed method is more effective and can provide a better indication for disease diagnosis than the baselines.
- Silver Standard

Conclusion
- A novel unsupervised deep learning framework to annotate phenotype from EHRs was proposed.
- The experiments in this work show the effectiveness and efficiency of the proposed method.
- Based on the results of the experiments, the proposed method achieved better performance and computational efficiency than other method, and can provide a better indication for disease diagnosis.
Future Work
Future work for this research includes:
- Integration of external biomedical literature from PubMed, Elsevier, etc.
- Annotation of all 13,000 specific phenotypes in HPO.
- Improvement of embedding of HPO by taking both semantics and hierarchy into consideration.
- Active learning strategies
- Application to general NLP domains
About Jingqing Zhang
Jingqing Zhang is a 3rd-year PhD (HiPEDS) at Department of Computing, Imperial College London under the supervision of Prof. Yi-Ke Guo. His research interest includes Natural Language Processing, Text Mining, Data Mining and Deep Learning. He received his BEng degree in Computer Science and Technology from Tsinghua University, 2016, and MRes degree with distinction in Computing from Imperial College London, 2017. In late 2019, he finished an internship with Brain Team, Google Research at California. Jingqing’s Masters and PhD at Imperial College, London have been funded via the HPCC Systems Academic Program, and he has used our big data analytics platform to process the data used in his research. He meets regularly with HPCC Systems team members who provide mentoring involving the use of HPCC Systems.
Acknowledgements
A special thank you to Jingqing Zhang for his phenomenal presentation, “Integrating Prior Knowledge with Learning in Biomedical Natural Language Processing.”
Additional Links
“Integrating Prior Knowledge with Learning in Biomedical Natural Language Processing”
Research at Data Science Institute, Imperial College London
Research at Data Science Institute, Imperial College London that leverages structured and unstructured data as prior knowledge, to improve deep learning models in natural language processing include the following:
Using Structured Data
- Integrating Semantic Knowledge to Tackle Zero-shot text Classification. NAACL 2019
- Unsupervised Annotation of Phenotypic Abnormalities via Semantic Latent Representations on Electronic Health Records. IEEE BIBM 2019.
Using Unstructured Data
- PEGASUS: Pre-training with Extracted Gap-sentences for Abstractive Summarization. Submitted to ICML 2020.
